Research
Below are some examples of current research at Columbia DBMI.
Large-Scale Evidence Generation and Evaluation in a Network of Databases (LEGEND)
|
The Observational Health Data Sciences and Informatics (OHDSI) consortium uses large-scale evidence to improve clinical practice. OHDSI announced the results of its Large-Scale Evidence Generation and Evaluation in a Network of Databases (LEGEND) study of hypertension. OHDSI researchers employed hundreds of millions of records from the OHDSI network and OHDSI’s state-of-the-art causal methods to address observed confounding and residual confounding to fill in the large gaps in evidence in the 2017 US hypertension guideline. The new guideline used randomized trial results wherever possible, but were forced to make many decisions based merely on expert opinion and to avoid many potentially helpful decisions. OHDSI tested over half a million hypotheses comparing 39 specific drugs, many more combinations of those drugs, and all classes of drugs on 58 outcomes related to efficacy and safety in the treatment of hypertension.
|
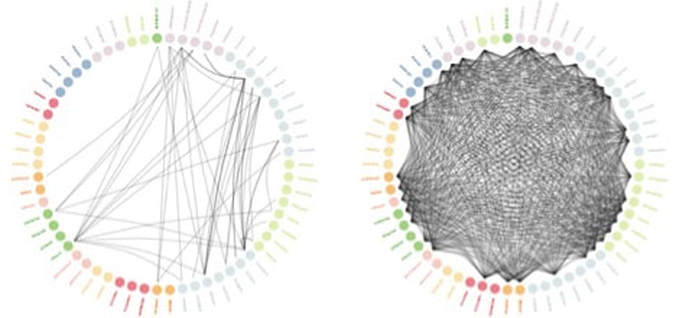
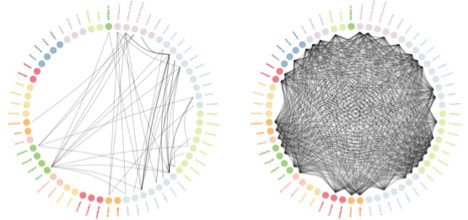
The left figure shows the available randomized trial evidence (circles represent individual drugs, and arcs represent studies that have compared those drugs), and the right figure shows the evidence added by OHDSI LEGEND.
|
|
Comparing OHDSI’s results to known randomized trial results, the two agreed on 28 out of 30 hypotheses that could be tested. This low rate of discordance may be similar to discordance among randomized trials.
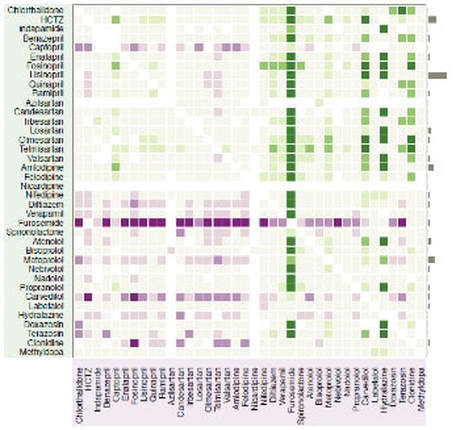
Cardiovascular outcomes effect sizes for single-drug comparisons are shown. On both axes, the guideline’s first-line drugs are listed first (i.e., top half of the y-axis and left half of the x-axis) and then second-line drugs. Green means the drug on the y-axis is better, and violet means the drug on the x-axis is better. The relative lack of color in the upper left quadrant shows minimal differences among first-line drugs, just as one would hope for the guideline’s recommendation. The green upper right and violet lower left quadrants imply that OHDSI finds first-line drugs superior to second-line drugs, again in keeping with the guideline. |
The general clinical lessons from the OHDSI study, which also included drug pairs and safety outcome measures, are as follows. OHDSI agrees with the US guideline that beta-blocker drugs are inferior to first-line drugs. This contradicts the European guideline, which marked beta blockers as first-line drugs. In contrast to the US guideline, OHDSI did detect one difference between two first-line drug classes, finding thiazide diuretics as superior to angiotensin-converting-enzyme inhibitors. The US guideline recommended beginning therapy with two drugs at once (based on expert opinion), whereas OHDSI found no difference in efficacy and worse safety for two drugs. One topic of clinical interest is related to two first-line drugs: whether chlorthalidone is superior to hydrochlorothiazide. OHDSI found no difference in efficacy and a possible difference in safety in favor of hydrochlorothiazide. This may inform a very large randomized trial that is currently in progress.
All the results of the OHDSI LEGEND hypertension study are freely available at data.ohdsi.org/LegendBasicViewer/. See OHDSI.org for more information about LEGEND and the protocol for this study, and see github.com/OHDSI for all the software related to this study.
All the results of the OHDSI LEGEND hypertension study are freely available at data.ohdsi.org/LegendBasicViewer/. See OHDSI.org for more information about LEGEND and the protocol for this study, and see github.com/OHDSI for all the software related to this study.
Human Phenotype Knowledge Discovery, Representation, and Application for Precision Medicine
|
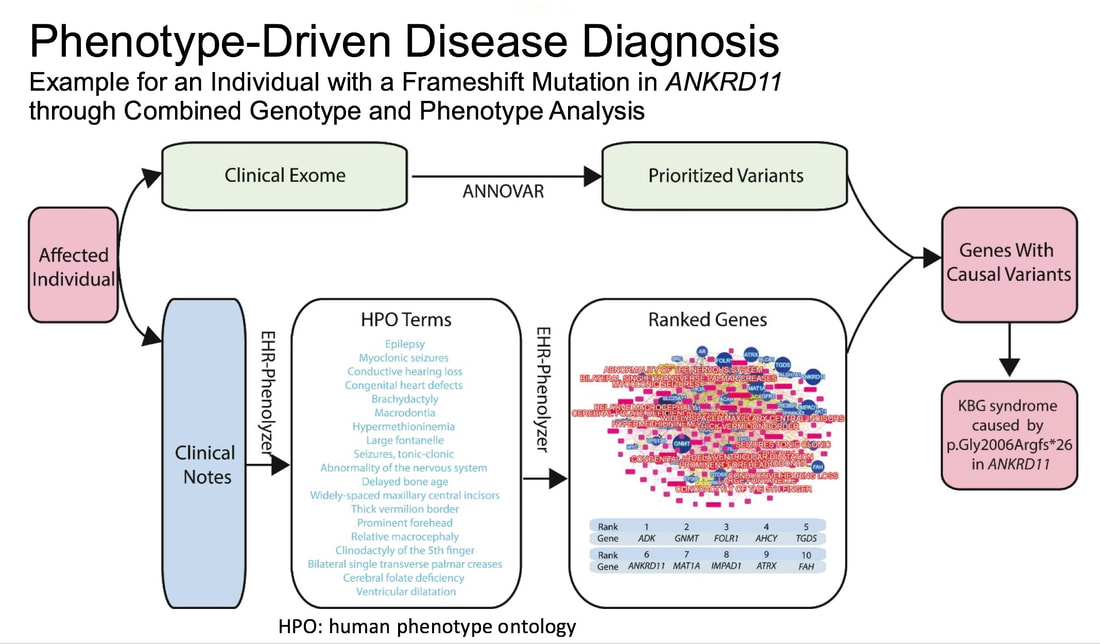
The Weng Lab develops informatics methods to extract characteristic phenotypes for genetic diseases from the electronic health records and normalize phenotype concepts using the Human Phenotype Ontology. The resulting phenotype can be used to study disease similarities towards a precise disease classification and to develop phenotype-driven diagnostic tools for patients affected by genetic diseases. The lab focuses on methodology development for phenotype knowledge engineering.
Son JH, Xie G, Yuan C, et al. Deep Phenotyping on Electronic Health Records Facilitates Genetic Diagnosis by Clinical Exomes. The American Journal of Human Genetics. 2018 Jul 5;103(1):58-73. |
Disease Heritability Inferred from Familial Relationships Reported in Medical Records
|
|
Heritability is essential for understanding the biological causes of disease, but requires laborious patient recruitment and phenotype ascertainment. Electronic health records (EHR) passively capture a wide range of clinically relevant data and provide a novel resource for studying the heritability of traits that are not typically accessible. EHRs contain next-of-kin information collected via patient emergency contact forms, but until now, these data have gone unused in research. Members of the Tatonetti Lab mined emergency contact data at three academic medical centers and identified millions of familial relationships while maintaining patient privacy. Identified relationships were consistent with genetically-derived relatedness. The team used EHR data to compute heritability estimates for 500 disease phenotypes. Overall, estimates were consistent with literature and between sites. Inconsistencies were indicative of limitations and opportunities unique to EHR research. These analyses provide a novel validation of the use of EHRs for genetics and disease research.
Polubriaginof FC, Vanguri R, Quinnies K, et al. Disease Heritability Inferred from Familial Relationships Reported in Medical Records. Cell. 2018 Jun 14;173(7):1692-704. |