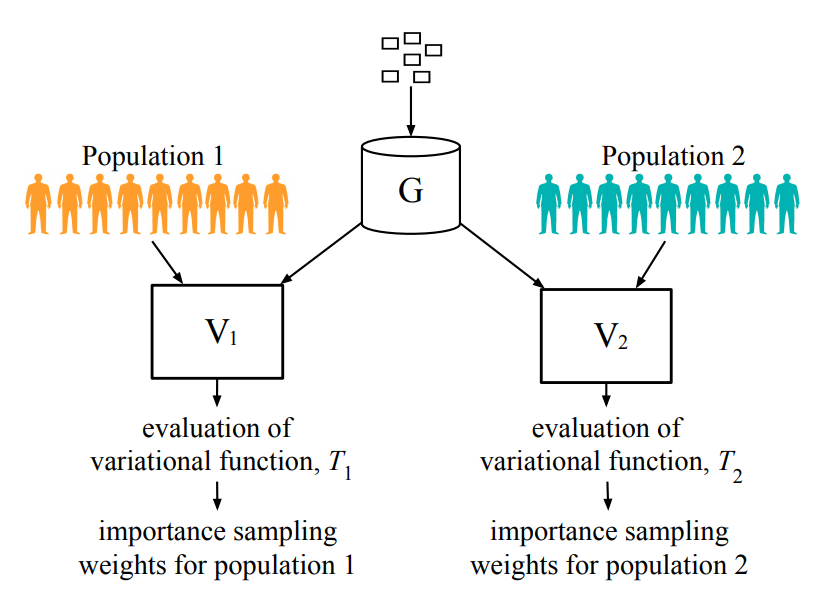
The Counterfactual χ-GAN
January, 2020
Averitt A, Vanitchanant, N, Ranganath R, Perotte A. "The Counterfactual χ-GAN." arXiv preprint arXiv:2001.03115. 2020 Jan 9. bib pdf

Averitt A, Vanitchanant, N, Ranganath R, Perotte A. "The Counterfactual χ-GAN." arXiv preprint arXiv:2001.03115. 2020 Jan 9. bib pdf
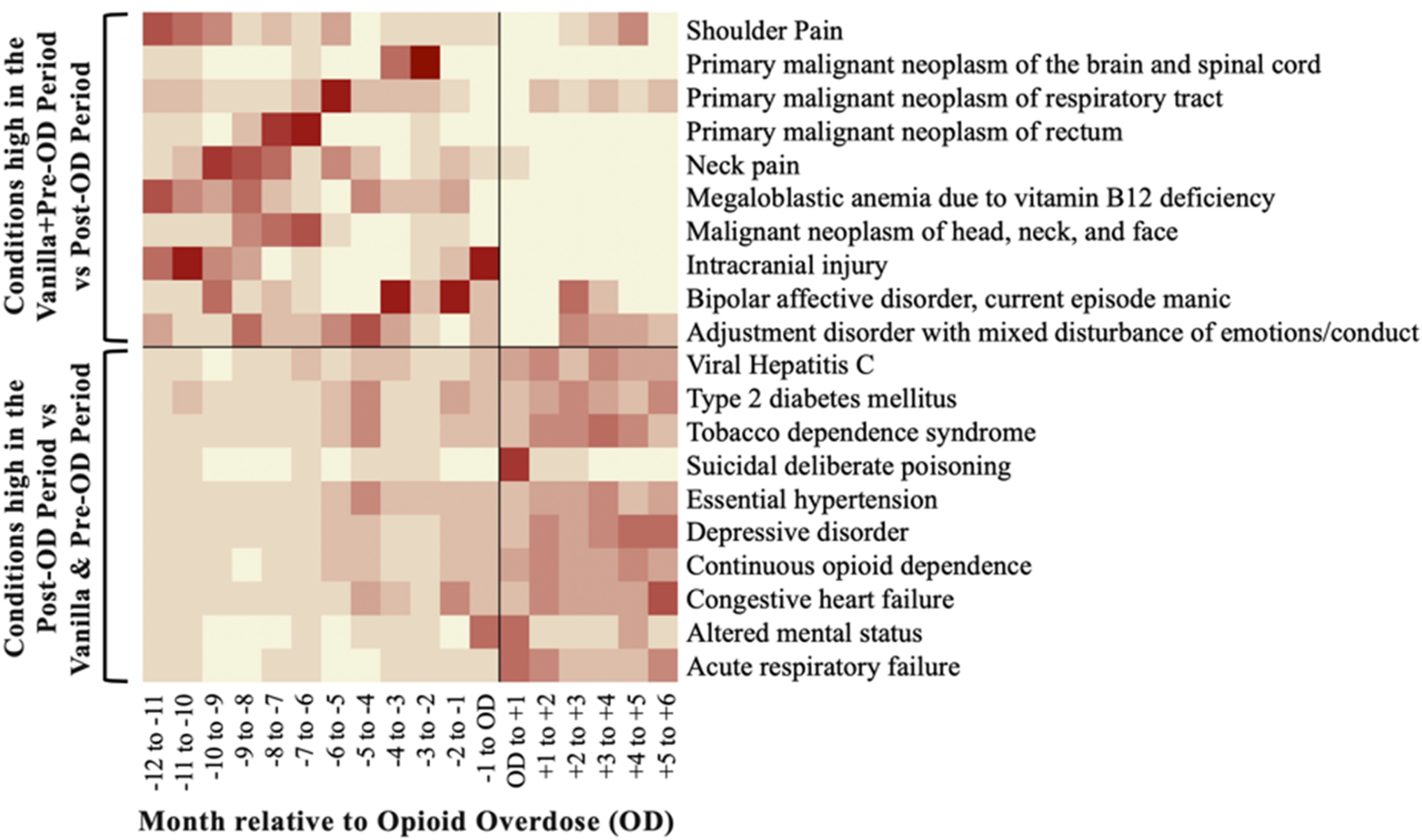
Averitt AJ, Slovis BH, Tariq AA, Vawdrey DK, Perotte AJ. Characterizing non-heroin opioid overdoses using electronic health records. JAMIA Open. 2019 Nov 26. bib pdf
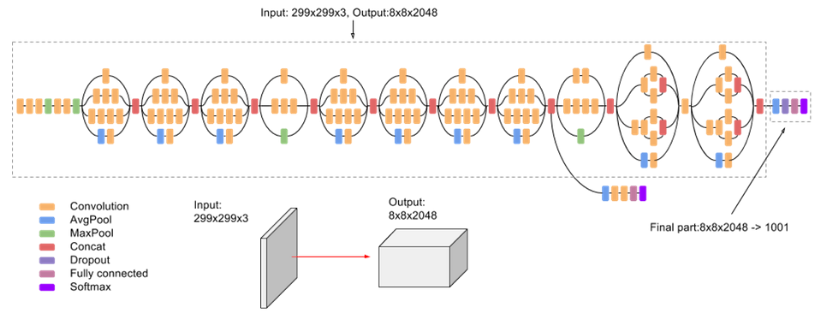
Pavinkurve NP, Natarajan K, Perotte AJ. Deep Vision: Learning to Identify Renal Disease With Neural Networks. Kidney International Reports. 2019 Jul;4(7):914. bib pdf
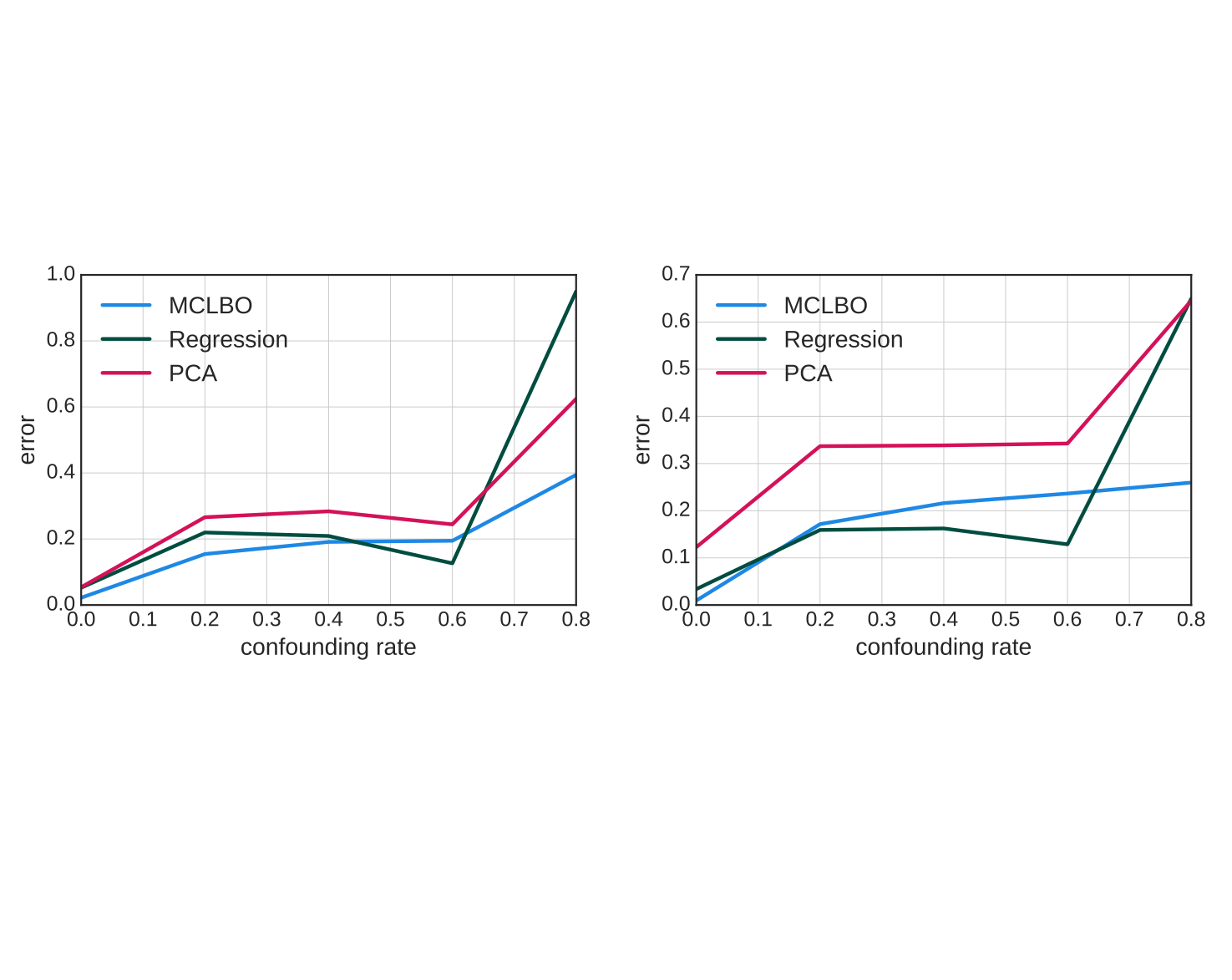
Ranganath R, Perotte A. "Multiple causal inference with latent confounding." arXiv preprint 2018. pdf
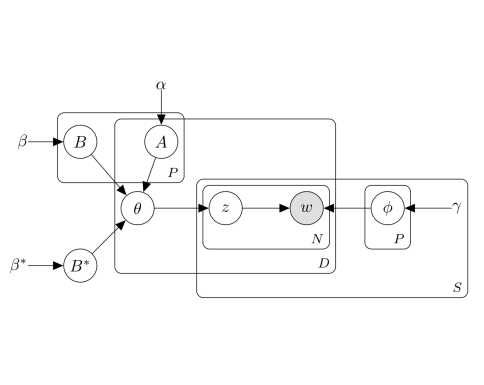
Rodriguez V, Perotte A. "Phenotype inference with Semi-Supervised Mixed Membership Models." NeurIPS Machine Learning for Health Workshop 2018 Dec 7. pdf
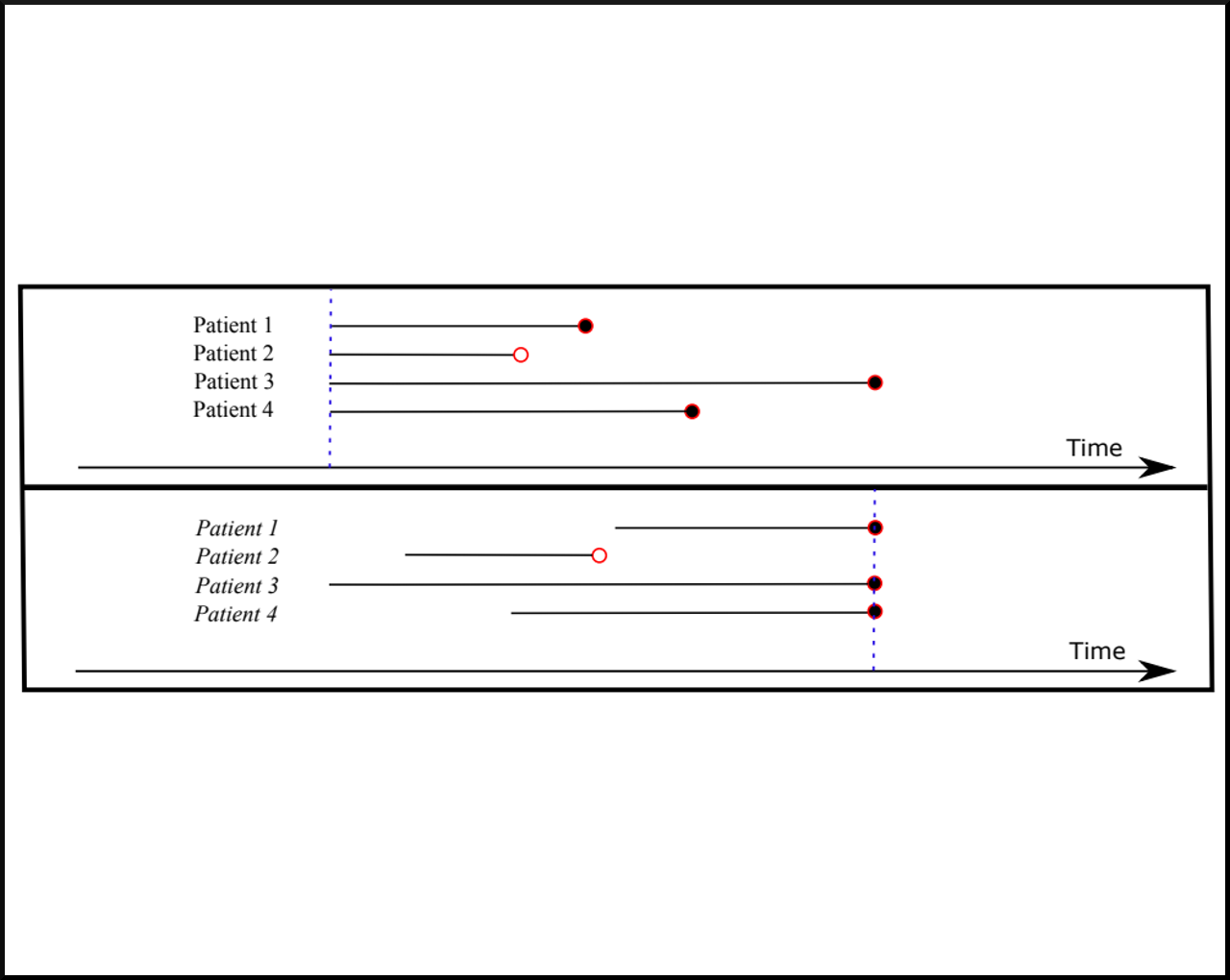
X Miscouridou, A Perotte, N Elhadad, R Ranganath. "Deep survival analysis: Missingness and Nonparametrics." In Machine Learning for Healthcare Conference, 244-256. 2018. bib pdf

I work on prediction and analysis of electronic health record data using existing and novel probabilistic methods.
I am interested in developing methods that combine disparate sources of data, such as clinical notes, laboratory values, medications, procedures, and billing codes to predict things such as chronic kidney disease progression or analyze data to identify side-effects that were previously unknown.
As an extention of my work in analyzing electronic health record data, I am also interested in developing new sources of data to improve our ability to predict and analyze.
I am interested in developing methods that leverage data from mass spectrometry and light spectroscopy to better characterize an individual's current state of health and predict their future state of health.
This course is targeted to biomedical scientists developing a broad understanding of computational methods applicable in biomedicine. This is a fast-paced, technical course covering a broad range of topics including: Density estimation, regression, classification, deep learning, probabilistic graphical models, clustering, dimensionality reduction, time series models, statistical NLP, networks, hypothesis testing, causal inference, imputation, and association rule mining. Students are expected to read technical texts carefully, participate actively in lecture discussion, and develop hands-on skills in labs involving real-world biomedical and health datasets.
This course is a reading course targeted towards biomedical scientists interested in developing in-depth knowledge of Bayesian statistics and the graphical modeling framework. This is a fast-paced course covering the fundamentals of probabilistic graphical modeling theory, exponential families, model design, latent variable models, the expectation maximization algorithm, and various methods for inference including Markov chain Monte Carlo methods and variational methods. This course will involve several programming assignments and a final project.
This course is targeted for biomedical scientists looking for working knowledge of programming and statistics. This is a fast-paced, hands-on course covering the following topics: programming basics in Python, probabilities, elements of linear algebra, elements of calculus, and elements of data analytics. Students are expected to learn lecture material outside of the classroom and focus on labs during class. All labs revolve around real-world biomedical and health datasets.
Weekly seminar series with invited speakers, student research talks, and journal clubs. View the seminar schedule here.
For a PDF version, please click here.
For links to my publications, please see my Google Scholar page.
Department of Biomedical Informatics
Columbia University Medical Center
622 West 168th Street. PH20
New York, New York 10032
http://people.dbmi.columbia.edu/~ajp9009/
Amelia Averitt - PhD Candidate
Shreyas Bhave - PhD Student
Victor Rodriguez - PhD Student
Katherine Schlosser - Masters
Aurnov Chattopadhyay - Undergraduate
Francesco Grechi - Undergraduate
Jason Ping - High School
Pierre Elias - Clinical Fellow
Peter Bullen - PhD Candidate, Applied Math and Applied Physics
Joongheum (PJ) Park - Clinical Informatics Fellow
Guillaume David - Postdoctoral Research Scientist
Anando Sen - Postdoctoral Research Scientist
Natnicha (Numfah) Vanitchanant - Masters
Liana Tascau - Masters
Aras Curukcu - High School Intern