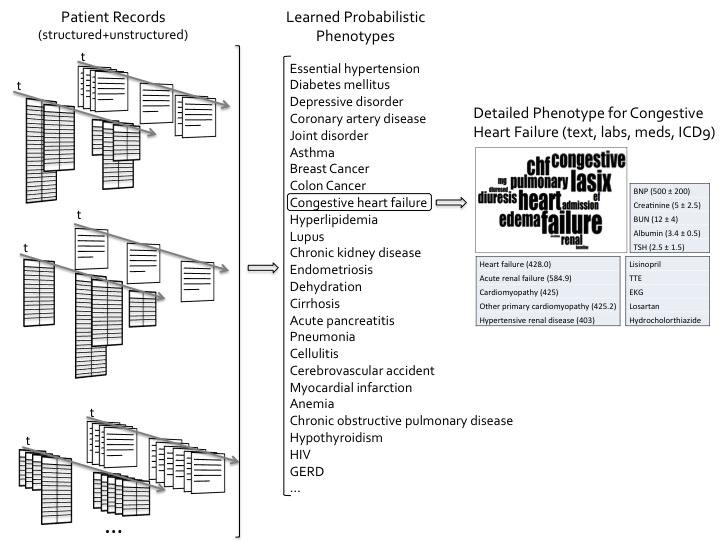
The research for this project contributes to two inter-related outcomes: (i) a probabilistic graphical model of a patient record and the patient's latent phenotypes. Models that can handle the heterogeneous data types in the EHR, along with their challenges, such as sparseness and artificial redundancy are investigated. For the models to be useful in the clinical world, they must be interpretable by humans, easily adaptable for EHR-driven applications, and clinically relevant. This is achieved by specifying prior clinical knowledge into the models and learning from clinicians' feedback automatically; and (ii) a patient record summarizer for clinicians at the point of patient care. HARVEST, our summarization system, leverages the probabilistic patient model and learns new models of salience through the clinicians' interactions with the deployed summarizer, in essence learning relevance of different patient phenotypes. For the evaluation of the phenome model and the summarizer, particular care is given to assessing their value in a real-world clinical setting.