Columbia University Department
of Biomedical Informatics
Discovering and Applying Knowledge in
Clinical Databases
The long term goal of our ongoing project, “Discovering and applying knowledge in clinical databases,” is to learn from data in the electronic health record (EHR) and to apply that knowledge to relevant problems. The increasing adoption of the EHR promises to provide data for clinical research and informatics research, but secondary use of the data has been limited. Challenges include the complexity, incompleteness, and inaccuracy of the record. Our current focus is to study the EHR from an information theoretic point of view, treating the EHR as a natural object worthy of study, and applying methods from non-linear time series analysis. Armed with a better understanding of the record, we hope to measure and account for data completeness and to improve interpretation and use of the data. We hypothesize that we can characterize an electronic health record using a formal information theoretic framework, and that the measured properties can help answer informatics and clinical questions.
The Team
|
|
George Hripcsak, MD, MS Vivian Beaumont Allen
Professor and Chair, Biomedical Informatics |
|
David Madigan, PhD Professor and Chair, Department
of Statistics |
|
|
Carol Friedman, PhD Professor of Biomedical
Informatics |
|
Colin Walsh, MD Postdoctoral fellow |
|
|
David Albers, PhD Associate Research Scientist of Biomedical Informatics |
|
|
Funded by a grant from the
National Library of Medicine, “Discovering and applying knowledge in clinical databases”
(R01 LM006910).
Current directions
1. Development of an
information theoretic approach to understanding electronic health record data
|
One of the
challenges of electronic health record data is that the data are sampled
irregularly, and usually when patients are ill, producing biased
retrospective experiments. We find that—after some lag—patients are sampled
more frequently when they are ill, and then the rate drops off as the disease
resolves. |
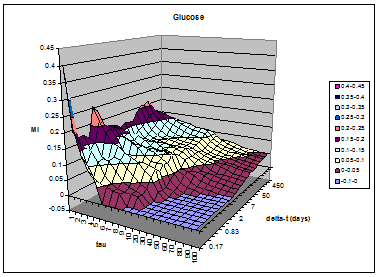
Here is a study of serum glucose, where
predictability—past values predicting a future value—is quantified as mutual
information. Tau is the number of glucose measurements between any pair of
values, and delta-t is the actual time between measurements. It reveals the
diurnal variation of glucose, as evidenced by the ridges at 24 hour
intervals. |
|
We have extended
non-linear time series analysis using mutual information to quantify
predictability, applying it to sparse and irregularly sampled time series. In
Chaos, Solitons & Fractals, we study its properties. |
Electronic health
records comprise the data of many patients. Aggregating across patients
brings its own challenges, which are presented in Chaos. |
2. Properties of electronic
health records and the effects of health care processes
|
In JAMIA, we studied parwise correlations in the
electronic health record using lagged linear correlation on a 3.7-million
patient, 24-year database. We found that there were several types of
associations: definitional associations included low blood potassium
preceding “hypokalemia”; low potassium preceding the drug spironolactone with
high potassium following spironolactone exemplified intentional and
physiologic associations, respectively. Counterintuitive results such as the
fact that diseases appeared to follow their effects may be due to the workflow
of healthcare, in which clinical findings precede the clinician’s diagnosis
of a disease even though the disease actually preceded the findings. |
In JAMIA, we discuss
the effects of health care processes on electronic health record data and
their likely effects on studies that use such data. We put forward a
framework for addressing these biases by studying the electronic health
record as an object of interest in itself and by creating a model of health
care processes. |
3. Population physiology
|
In PLOS ONE, we studied
glucose control in populations of patients using electronic health record
data. We showed that we could validate a physiologic
glucose model using time-delayed mutual information even though the raw data
values had too much noise for the same task. |
In Annals
of Neurology, we study a population of patients with subarachnoid hemorrhage to
better understand whether secondary seizures cause further damage or are
simply harmless passengers. Seizures do in fact appear to be associated with
outcomes. |
|
In Physics Letters A, we
demonstrate our statistical dynamics approach to study physiology at the
population scale. We confirm diurnal variation in creatinine
data. |
|